ABOUT OUR PRODUCT
While DNA sequencing is an integral part of genetics research, there is a significant lack of fully-functional tools that provide easy access to DNA sequencing in a cost-effective manner.
Our goal is to minimize the inefficiencies faced by professionals who conduct NGS for Chloroplast, Mitochondria and plant ITS regions by developing a complete solution that incorporates existing DNA sequencing pipelines into a single environment that is both intuitive and accessible.
- Provides a GUI environment integrating all the necessary tools for the pipelines.
- Automates many of the intermediate steps while providing the user the relevant information and statistics for each step.
- Provides suggestions to the user on possible next steps to take in case there are multiple possibilities.
Background
Our product is focused on the domain of Next Generation Sequencing and Skim sequencing technique.
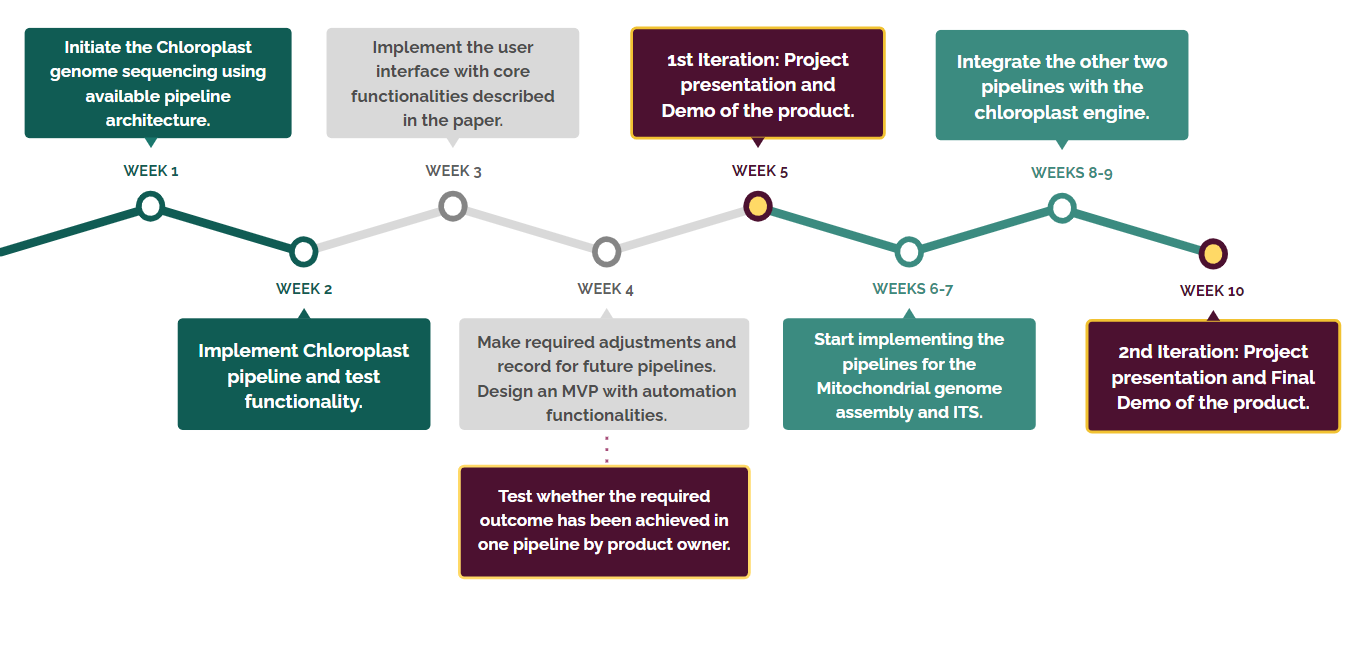
Timeline
OUR OBJECTIVE
Our goal is to minimize the inefficiencies and problems faced by the professionals who conduct their research in NGS for Chloroplast, Mitochondria and plant ITS regions. We plan to provide this combined functionality of each stage in the process of NGS, through a well-planned combination of pipelines, in the final form of an open source software with additional features which will provide the users the opportunity to carry out NGS with ease, so that they can make decisions while receiving recommendations made by us in order to achieve maximum accuracy required for the process.
Higher efficiency
To minimize the inefficiencies and problems faced by the researchers, while optimizing the processing time through multi-threading.
Combined functionality
Provide this combined functionality of each stage in the process of NGS, through a well-planned combination of pipelines.
Open-source
Provide an open source software with additional features at each step of the pipeline.
Increased performance
Provide a platform for users to carry out Skim sequencing without having to manually choose the tools with best performance.
Single package
Provide a single package with dependencies and resolve issues with different file formats.
Recommendations
Additional functionalities include providing recommendations to the user in order to improve performance. This is handled by an ML component.
Process
The users can select from the three pipelines; Chloroplast, Mitochondria and ITS region and select the an assembly tool of their choice, among the ones that have been provided by us in the toolkit. The running of the process is automated along with many of the configuration decisions that need to be set prior to running. The users can observe the relevant output logs once the assembly process is over, along with the annotated genomes and necessary statistics.
Integrate the pipelines
The solution will contain a GUI environment that integrates all the necessary tools for the pipelines into a single space.
Automate the Workflow
The software will automate a lot of the intermediate steps while providing the user the relevant information and statistics for each step.
Provide Recommendations
The solution will attempt to provide suggestions to the user on possible next steps to take in case there are multiple possibilities.

Workflow
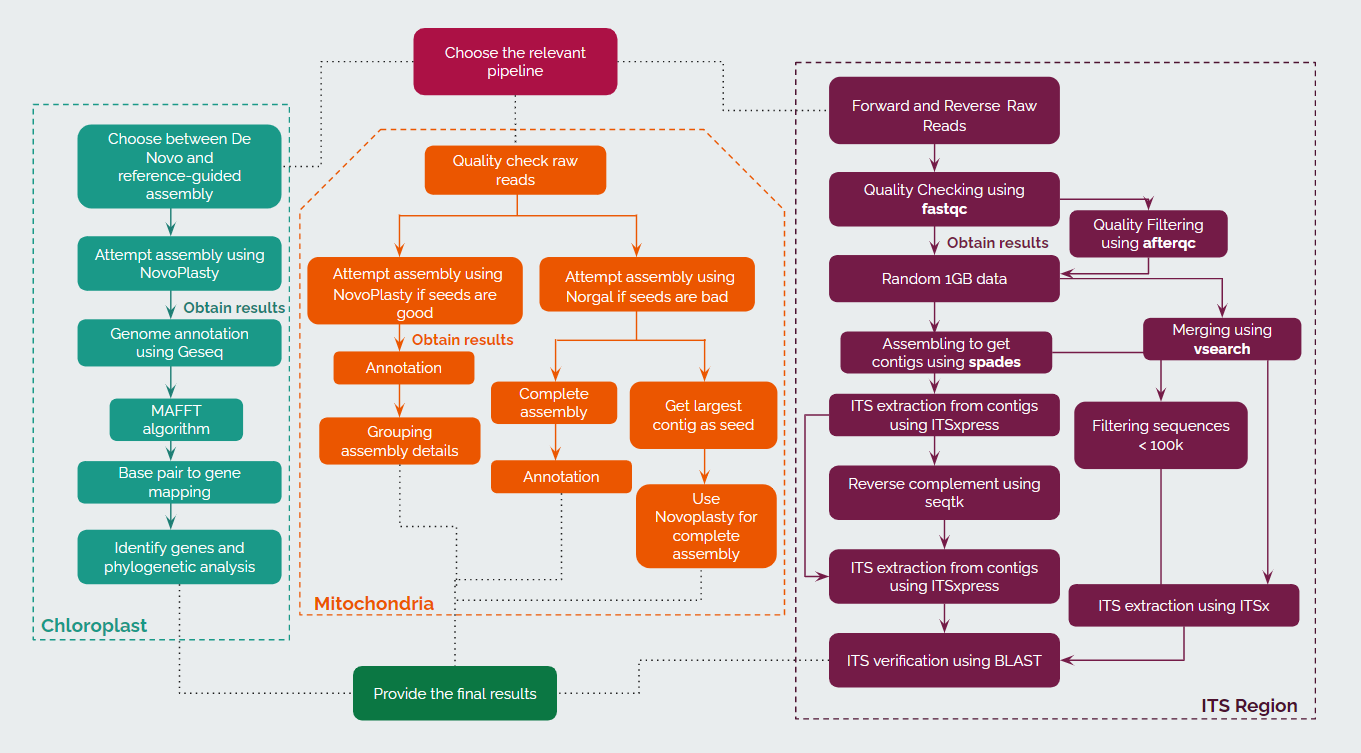
In the general workflow of the system, the user can choose among the existing pipelines for Chloroplast, Mitochondria and ITS regions. The steps under the implementation of each pipeline are given above. We will be using many open-source assembly tools such as FastPlast, NovoPlasty and GetOrganelle, along with other tools for pre-processing of data and for annotations. The outputs provided by these tools will be presented to the user at each step of the pipeline.
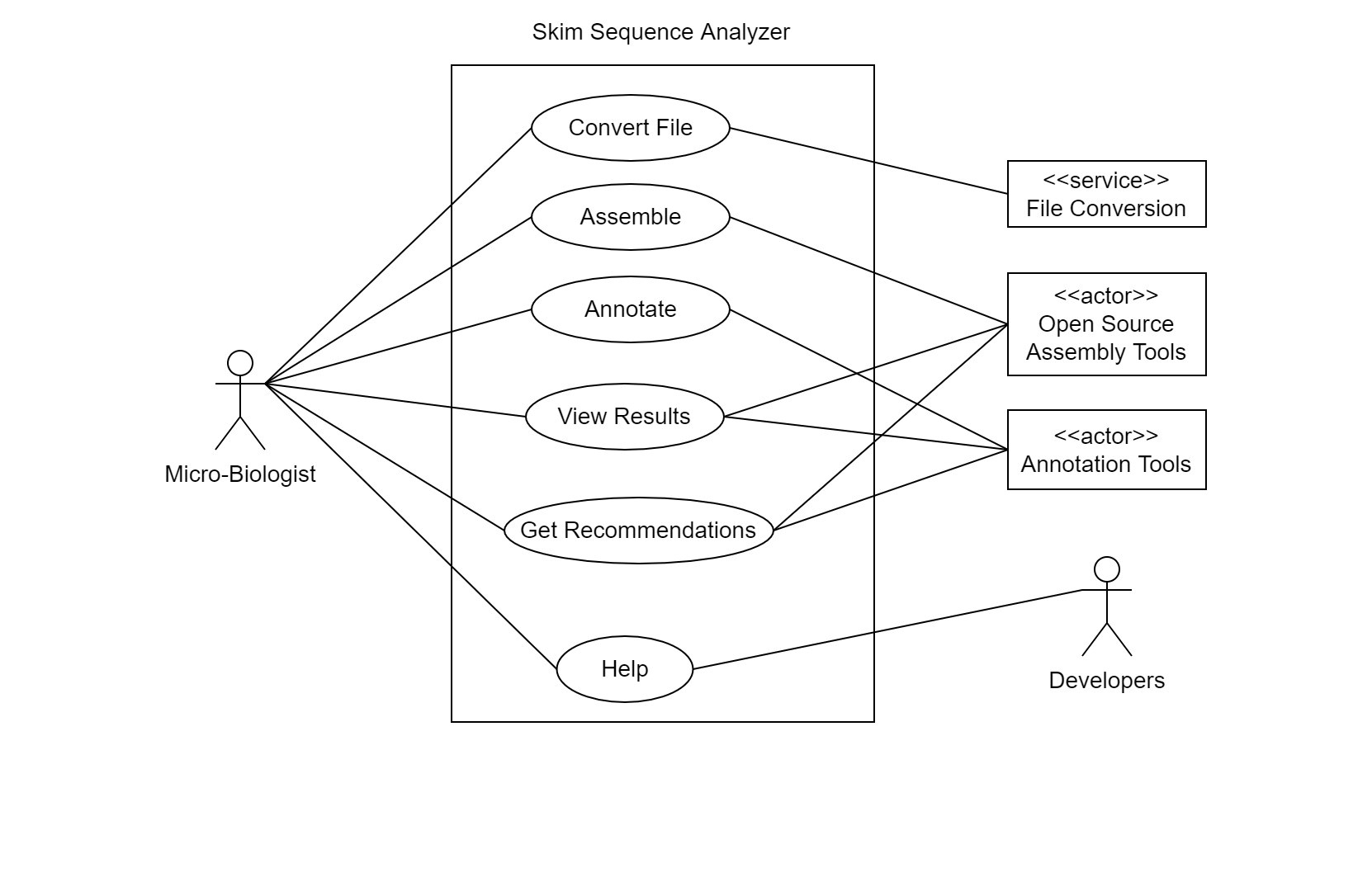
UML Diagrams
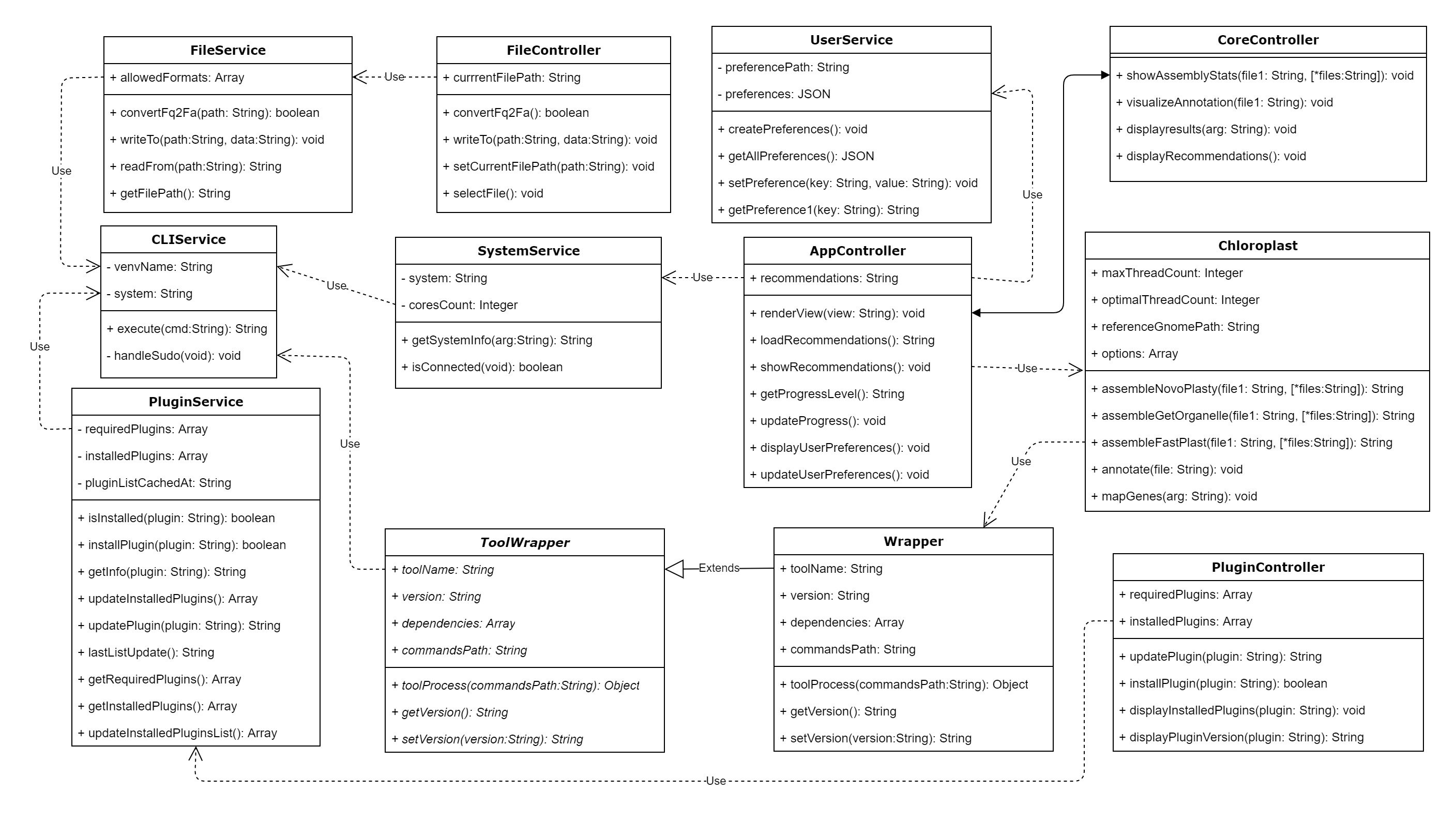
Software Architecture
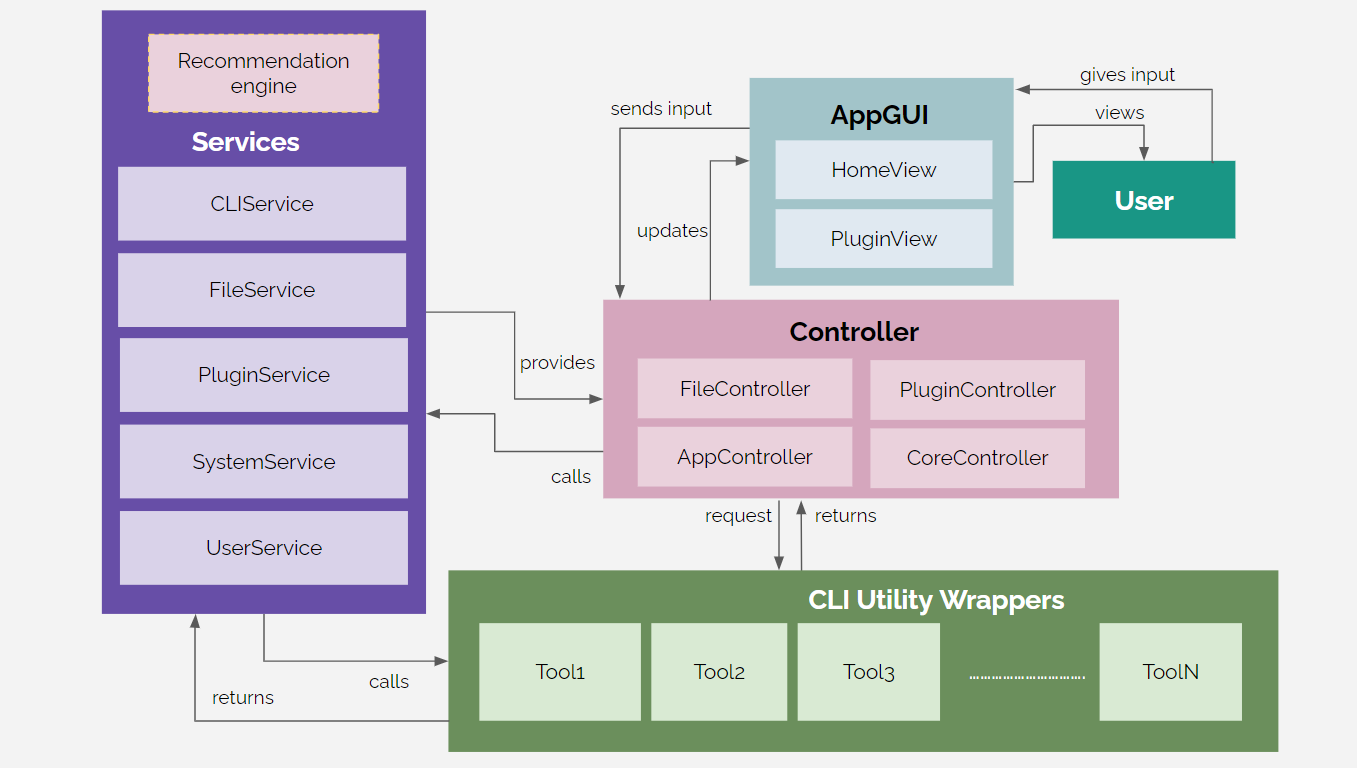
We have used the MVC architecture for the development our software. The User will be directly providing input through the App GUI, which displays all the necessary information to the user. The GUI is directly updated through the Controllers. There are several Controllers which associate with the methods defined in the Services and the Tool Wrappers in order to provide the pipeline functionality. There are multiple Services in the system to handle CLI commands, file-format handling, dependency installation, storing user preferences, check connectivity, get necessary file path for input files, and many more. All the open-source tools we are using for implementing the pipelines will be handled using Tool Wrappers for the ease of implementation. The recommendation engine will be running as a seperate part of the Services provided by the software.
GUI Mockup
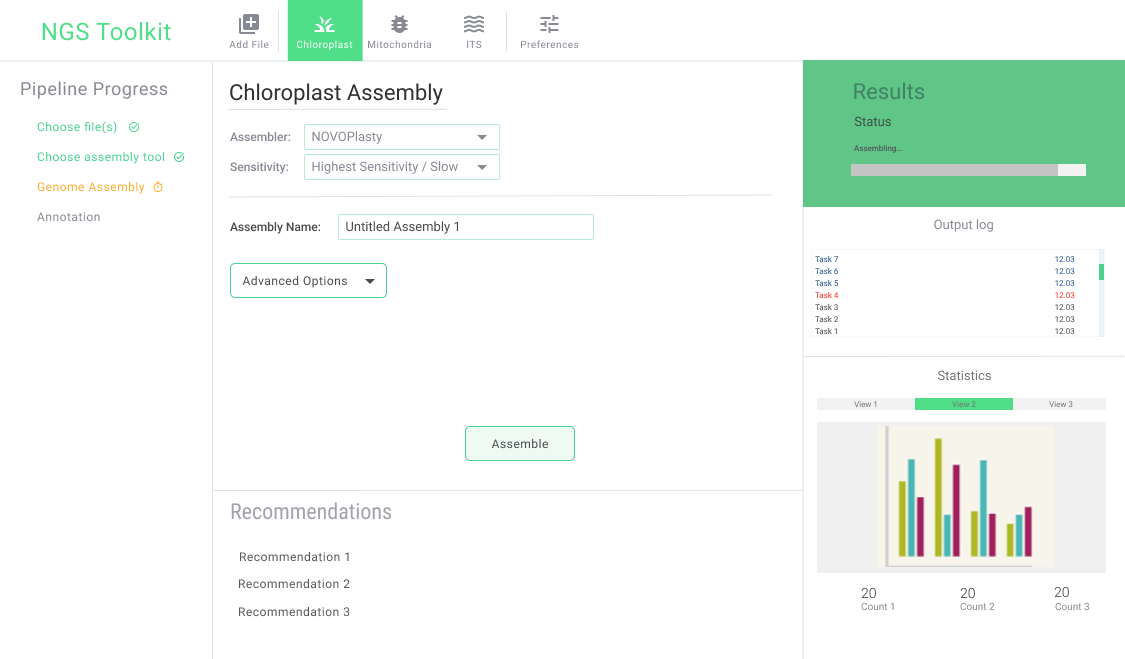
Current Progress
Given below is the GUI that our team is currently working on.
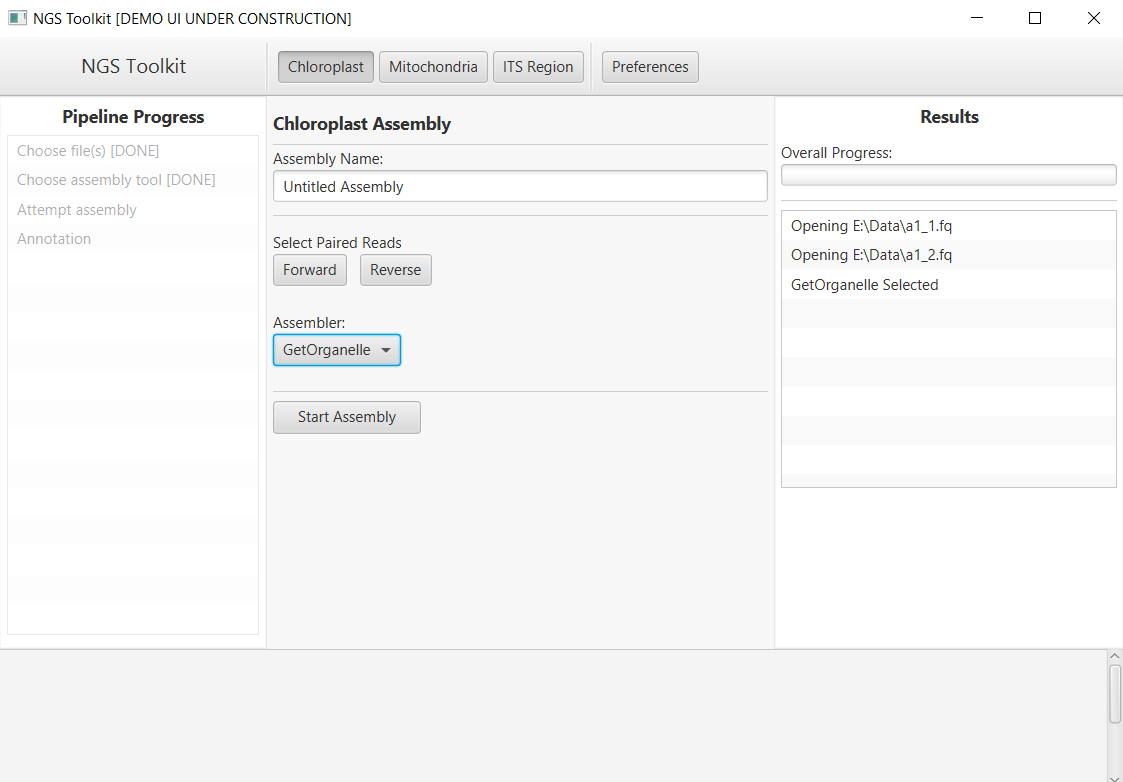
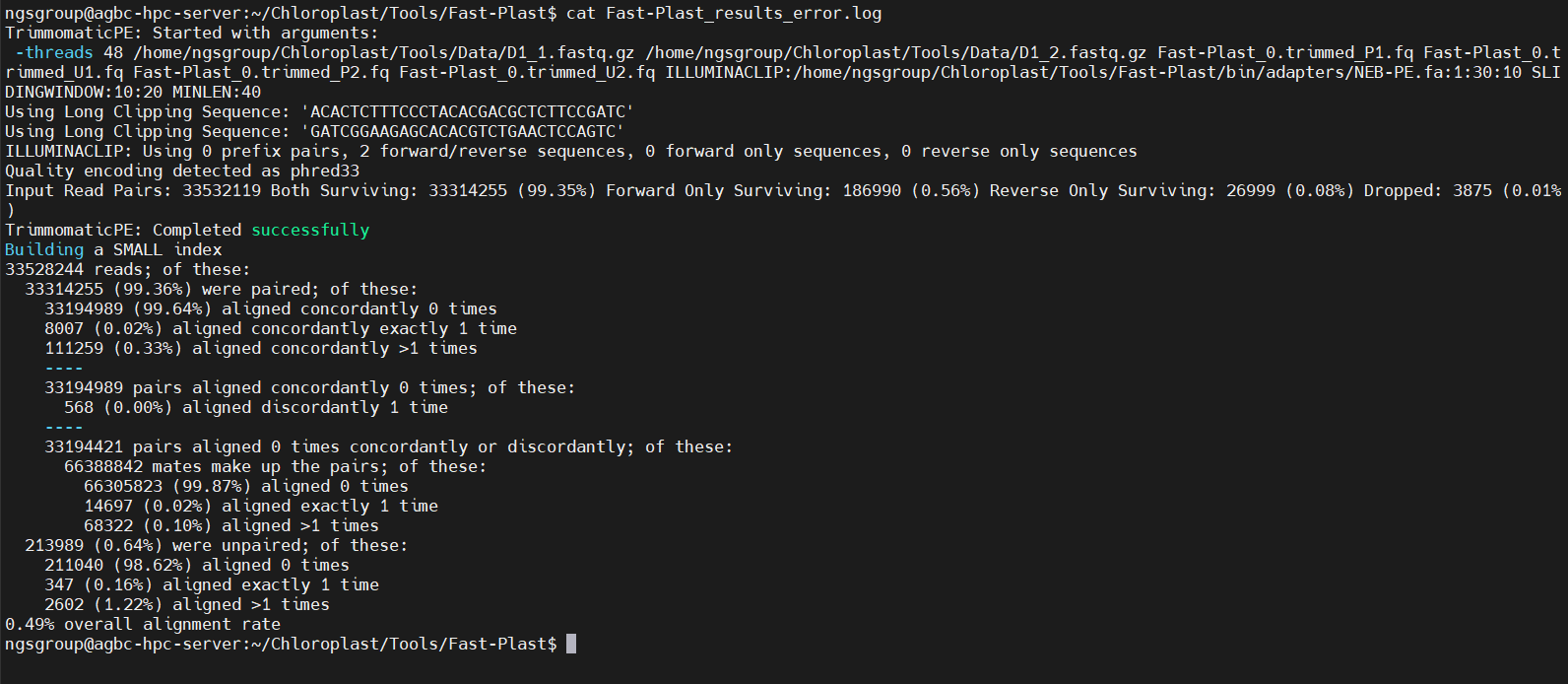
The implementation of the Fast-Plast tool in the server and the obtained results for selected input files are shown below.